Euihyun Lee(이의현, 박사과정, 제1저자)
Fluorescent proteins contain an internal chromophore constituted of amino acids or an external chromophore
covalently bonded to the protein. To increase their fluorescence intensities, many research groups have
attempted to mutate amino acids within or near the chromophore. Recently, a new type of fluorescent
protein, called UnaG, in which the ligand binds to the protein through many noncovalent interactions was
discovered. Later, a series of mutants of the UnaG protein were introduced, which include eUnaG with
valine 2 mutated to leucine emitting significantly stronger fluorescence than the wild type and V2T mutant,
in which valine 2 is mutated to threonine, emitting weaker fluorescence than the wild type. Interestingly,
the single mutation sites of both eUnaG and V2T mutants are distant from the fluorophore, bilirubin, which
renders the mechanism of such fluorescence enhancement or reduction unclear. To elucidate the origin of
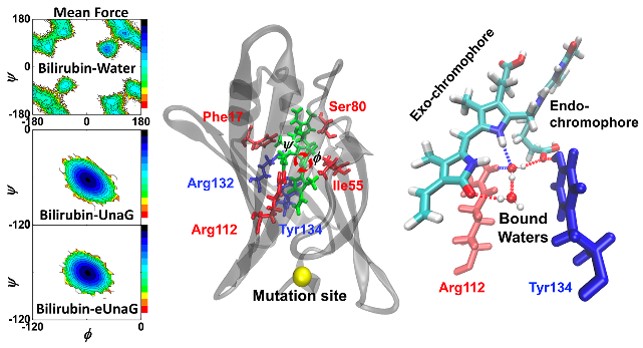
fluorescence intensity changes induced by the single mutations, we carried out extensive analyses on MD
simulations for the original UnaG, eUnaG and V2T, and found that the bilirubin ligand bound to eUnaG is
conformationally more rigid than the wild-type, particularly in the skeletal dihedral angles, possibly resulting
in the increase of quantum yield through a reduction of non-radiative decay. On the other hand, the
bilirubin bound to the V2T appears to be flexible than that in the UnaG. Furthermore, examining the
structural correlations between the ligand and proteins, we found evidence that the bilirubin ligand is
encapsulated in different environments composed of protein residues and water molecules that increase or
decrease the stability of the ligand. The changed protein stability affects the mobility and confinement of
water molecules captured between bilirubin and the protein. Since the flexible ligand contains multiple
hydrogen bond (H-bond) donors and acceptors, the H-bonding structure and dynamics of bound water
molecules are highly correlated with the rigidity of the bound ligand. Our results suggest that, to
understand the fluorescence properties of protein mutants, especially the ones with noncovalently bound
fluorophores with internal rotations, the interaction network among protein residues, ligand, and water
molecules within the binding cavity should be investigated rather than focusing on the local structure near
the fluorescing moiety. Our in-depth simulation study may offer a foundation for the design principles for
engineering this new class of fluorescent proteins.
This article is part of the themed collection: 2018 Chemical Science HOT Article Collection
https://pubs.rsc.org/en/Content/ArticleLanding/2018/SC/C8SC03558J#!divAbstract
 Topotactic Transformations in an Icosahedral Nanocrystal to F...
Topotactic Transformations in an Icosahedral Nanocrystal to F...
 Elucidating the Role of Molecule-Electrode Interfacial Defect...
Elucidating the Role of Molecule-Electrode Interfacial Defect...